After skipping a day, you may be expecting something special, and while I do think this is special, it's also a little focused. I make no apologies (and don't think I'm promising a new big post every day, although I'm trying). The subject this time is a very high-energy type of cellular "signaling" molecule: pyrophosphorylated inositols.
Inositols
Let's start with inositol: a sugar (sort of) that plays an important role in the cell metabolism.

Figure 1: Chemical structure of inositol. (From Wiki)
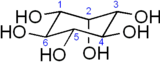
Figure 2: 3D Structure of inositol. (From Wiki)
Like most sugars, it has about one hydroxyl group per carbon atom, although "true" sugars like glucose actually have one more (or rather an aldehyde, which can and usually does hydrolyse to two hydroxides then dehydrolyse to produce the ring usually seen in sugars). Note that the inositol ring doesn't contain an oxygen like most sugars, it's a complete carbon ring.
Inositol comes in a number of isomers, differing in how they are organized around the carbon atoms in the ring.
Pyrophosphates
I've already discussed some of inositol's phospates as signaling molecules, but those were simple phosphates. A pyrophosphate is two phosphates hooked in a row, as in Adenosine diphosphate.
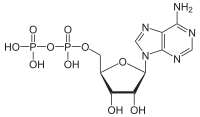
Figure 3: Chemical formula of adenosine diphosphate. Note the two phosphate groups in a row, with the very high-energy P-O-P bond between them. (From Wiki)
Inositol comes in a variety of phosphates.

Figure 3: Inositol with five carbons phosphorylated and one carbon (1, at the bottom) pyrophosphorylated. (From Nature Chemical Biology.)
The ones we're concerned with are those having one or more pyrophosphate groups, as well as phosphates. Some of these molecules have every carbon at least phosphorylated, others don't.
As with protein phosphorylation, these phosphate groups appear to derive from the third phosphate of Adenosine Triphosphate (ATP). The energy of the "P-O-P" bond in inositol pyrophosphates is intrinsically much greater than that of a "C-O-P" bond, roughly the same as the "P-O-P" bond in ATP. This means the reaction may well proceed somewhat more slowly. However, the situation within the cell is highly confusing, including substantial concentrations of Magnesium (Mg++), which can complex with the negatively charged phosphate groups, potentially making the bond more favorable. From one study:2
The standard free energy of hydrolysis of the pyrophosphate bond in IP7 has been estimated theoretically for the nonphysiological isomer 1PP-IP5 at 6.6 kcal/mol, higher than that of adenosine 5'-diphosphate (ADP) (6.4 kcal/mol) and lower than that of adenosine 5'-triphosphate (ATP) (7.3 kcal/mol) (1).
When it comes to dephosphorylation, no sign has yet been found that it is used as a short term signal:6
Since the DIPPs [“Diphosphoinositol Polyphosphate Phosphohydrolases”, that is enzymes that dephosphorylate inositol pyrophosphates] are so active, their acute spatiotemporal regulation could greatly impact upon the turnover of the diphosphoinositol polyphosphates. However, there is as yet no evidence that this is exploited as a short-term signaling mechanism; no covalent modification of DIPPs has been found ([ref]). Furthermore, studies with GFP-DIPP constructs offer no evidence of their compartmentalization; the fusion proteins were uniformly distributed throughout the cell ([refs]). As for the intracellular distribution of endogenous DIPPs, we do not currently have antibodies with sufficient sensitivity for such studies.
It is possible that pyrophosphatase activity could split the entire pyrophosphate from the inositol molecule,2 although no evidence seems to have yet been found for this. Nevertheless, it is an intriguing possibility: the energy to be gained from splitting a pyrophosphate group free would be quite high due to the extremely low concentration of inorganic pyrophosphate (PPi) in the cytoplasm.
Two new, important papers
Thus to the first of two papers discussed here:6
Shears, S. (2009). Diphosphoinositol Polyphosphates: Metabolic Messengers? Molecular Pharmacology DOI: 10.1124/mol.109.055897
In general, inositol pyrophosphates are ideal in several ways as signaling (computing) intermediates:
The inositol phosphate family represents the convergence of several “good signaling ideas”, most notably in their use of a recurring theme in the cell-signaling genre, namely, phosphate groups. Phosphates have two especially prominent features that facilitate specificity of interactions between cell signaling entities. First, the bulky nature of the phosphate group imposes geometric constraints on ligand-protein and protein-protein interactions. Second, the phosphate’s negative charge at physiological pH bestows specificity on its interactions with target proteins through multiple ionic and hydrogen bonds. The negative charges on the phosphate group also make soluble, phosphorylated molecules lipid-impermeant, so that they can be retained inside cells.
Inositol offers several additional assets for a signaling molecule. It is chemically stable, it is small (hence it diffuses through cytosol quickly), and it is only a short synthetic offshoot from the glycolytic pathway ([ref]). There is also a functionally significant plane of symmetry across the 2/5-axis of the inositol ring ([Figure]). That symmetry permits one inositol phosphate to imitate another’s three-dimensional phosphate recognition pattern, when the orientation of the inositol ring changes in relation to the protein’s ligand-binding site ([ref]), although in addition the binding site itself has to be somewhat flexible ([ref]). This phenomenon provides a molecular explanation for the metabolic promiscuity of certain inositol phosphate kinases ([ref]) and it also facilitates functionally important metabolic interactions between different inositol phosphates ([ref]). With all of these advantageous features, it is not surprising that the inositol phosphates are a phylogenetically-widespread family of second messengers.
The disadvantage is pretty much the same as enzyme phosphorylation, it costs one ATP for each molecule phosphorylated. If, as I suspect, there are pathways that involve splitting a pyrophosphate group directly off the sugar, this would cost two ATPs.
Now, for the second paper to be discussed:1
Losito, O., Szijgyarto, Z., Resnick, A., & Saiardi, A. (2009). Inositol Pyrophosphates and Their Unique Metabolic Complexity: Analysis by Gel Electrophoresis PLoS ONE, 4 (5) DOI: 10.1371/journal.pone.0005580
This paper discusses "a novel, sensitive and rapid polyacrylamide gel electrophoresis (PAGE)-based method for the analysis of inositol pyrophosphates." It appears to be both much faster, and much more accurate, allowing more effective study of how these molecules behave in vitro, and the research reported here revealed:1
[...] the existence of a number of additional, previously uncharacterised pyrophosphorylated inositol species. More importantly, parallel analyses comparing SAX-HPLC- and PAGE-based methods reveal a significant underestimation of the quantity and composition of inositol pyrophosphate metabolism. Inositol pyrophosphates are typically subjected to acidic conditions before and during SAX-HPLC analysis. The study of these treatments using PAGE revealed the degrading effects of such actions.
In other words, not only are inositol pyrophosphates ideal mediators of internal cellular calculations (signaling molecules), but there appear to be many more of them in play than anybody had realized.
Implications
Now, let's move on in a more speculative vein.
While the energy involved in pyrophosphorylating inositols may not be that great, the energy involved in either breaking the "P-O-P" bond or splitting the entire pyrophosphate group off the sugar is quite high, especially the latter. This means that a "signaling" molecule could potentially adjust the local concentration of any particular pyrophosphate about as quickly as diffusion could bring them to it. The fact that no known phosphatase that affects pyrophosphorylated inosital has been found to be subject to covalent modifications (see above)6 doesn't mean they can't participate in analog computation: it may well be that covalent modifications of other, as yet undiscovered, enzymes change how they cluster, modifying the phosphatase (or pyrophosphatase) activity. For that matter, in budding yeast, increased concentration of a certain pyrophosphorylated inositol "interacts noncovalently with Pho80-Pho85-Pho81 and induces additional interactions between Pho81 and Pho80-Pho85 that prevent substrates from accessing the kinase active site. [my emphasis]"3 This concentration increase appears to be associated with phosphate starvation.4
The possibility that these molecules could be both involved in modifying high-speed phosphatase activity and also targets of that activity is quite intriguing. The very high energies involved in the reactions could potentially drive localized calculations at very high speeds. The fact that there are so many different species means that they could be driving changes to one another's concentrations in quite complex ways. Pyrophosphorylated Inositols have been implicated in "a wide range of biological functions, including vesicular trafficking, apoptosis, DNA repair, telomere maintenance, and stress responses [ref]. Similarly, important human diseases such cancer and diabetes appear to be under inositol pyrophosphate control [ref]."1 Many of these processes potentially require very localized, short-term control of the sort that these molecules could provide.
All in all, there's a great deal of research waiting to be done, and the new method documented in Losito et al.1 will hopefully play a major part. However, equally new, powerful, methods will have to be developed to allow much more precise in vivo research, more localized in both space and time.
Links: (These aren't in any particular order. Use the back key to return if you clicked on a footnote.)
1. Inositol Pyrophosphates and Their Unique Metabolic Complexity: Analysis by Gel Electrophoresis
2. Phosphorylation of Proteins by Inositol Pyrophosphates (Requires free registration)
3. Molecular basis of cyclin-CDK-CKI regulation by reversible binding of an inositol pyrophosphate
4. Regulation of a Cyclin-CDK-CDK Inhibitor Complex by Inositol Pyrophosphates (Requires free registration)
5. Protein pyrophosphorylation by inositol pyrophosphates is a posttranslational event
6. Diphosphoinositol Polyphosphates: Metabolic Messengers? (Pre-print, version 2)

Inositol pyrophosphates are recognized components of cellular processes that regulate vesicle trafficking, telomere length, and apoptosis, it is recently characterized cell signalling molecules responsible for the pyrophosphorylation of protein substrates. Also inositol pyrophosphates is intrinsically much greater than that of a "C-O-P" bond. Thanks a lot!
ReplyDelete